| 中文名稱 | DNA補充修復XPC細胞蛋白抗體 |
| 別 名 | Xeroderma pigmentosum complementation group C; DNA repair protein complementing XP C cells; DNA repair protein complementing XP-C cells; DNA repair protein complementing XPC cells; p125; RAD4; Xeroderma pigmentosum group C complementing protein; Xeroderma pigmentosum group C protein; Xeroderma pigmentosum group C-complementing protein; Xeroderma pigmentosum group III; XP 3; XP C; XP group C; XP3; Xpc; XPC gene; XPC_HUMAN; XPCC. |
| 研究領域 | 細胞生物 表觀遺傳學 |
| 抗體來源 | Rabbit |
| 克隆類型 | Polyclonal |
| 交叉反應 | Human, Mouse, Rat, (predicted: Dog, Pig, Cow, Horse, Guinea Pig, ) |
| 產品應用 | ELISA=1:500-1000 IHC-P=1:100-500 IHC-F=1:100-500 IF=1:100-500 (石蠟切片需做抗原修復) not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. |
| 分 子 量 | 106kDa |
| 細胞定位 | 細胞核 細胞漿 |
| 性 狀 | Liquid |
| 濃 度 | 1mg/ml |
| 免 疫 原 | KLH conjugated synthetic peptide derived from human Xeroderma pigmentosum group C:841-940/940 |
| 亞 型 | IgG |
| 純化方法 | affinity purified by Protein A |
| 儲 存 液 | 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. |
| 保存條件 | Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. |
| PubMed | PubMed |
| 產品介紹 | The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER; it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1. Function: Involved in global genome nucleotide excision repair (GG-NER) by acting as damage sensing and DNA-binding factor component of the XPC complex. Has only a low DNA repair activity by itself which is stimulated by RAD23B and RAD23A. Has a preference to bind DNA containing a short single-stranded segment but not to damaged oligonucleotides. This feature is proposed to be related to a dynamic sensor function: XPC can rapidly screen duplex DNA for non-hydrogen-bonded bases by forming a transient nucleoprotein intermediate complex which matures into a stable recognition complex through an intrinsic single-stranded DNA-binding activity. The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER; it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1. Subunit: Component of the XPC complex composed of XPC, RAD23B and CETN2. Interacts with RAD23A; the interaction is suggesting the existence of a functional equivalent variant XPC complex. Interacts with TDG; the interaction is demonstrated using the XPC:RAD23B dimer. Interacts with SMUG1; the interaction is demonstrated using the XPC:RAD23B dimer. Interacts with DDB2. Interacts with CCNH, GTF2H1 and ERCC3. Subcellular Location: Nucleus. Cytoplasm. Note=Omnipresent in the nucleus and consistently associates with and dissociates from DNA in the absence of DNA damage. Continuously shuttles between the cytoplasm and the nucleus, which is impeded by the presence of NER lesions. DISEASE: Defects in XPC are a cause of xeroderma pigmentosum complementation group C (XP-C) [MIM:278720]; also known as xeroderma pigmentosum III (XP3). XP-C is a rare human autosomal recessive disease characterized by solar sensitivity, high predisposition for developing cancers on areas exposed to sunlight and, in some cases, neurological abnormalities. Similarity: Belongs to the XPC family. SWISS: Q01831 Gene ID: 7508 Database links: Entrez Gene: 7508 Human Entrez Gene: 22591 Mouse Entrez Gene: 312560 Rat Omim: 613208 Human SwissProt: Q01831 Human SwissProt: P51612 Mouse Unigene: 475538 Human Unigene: 2806 Mouse Unigene: 22820 Rat Important Note: This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |
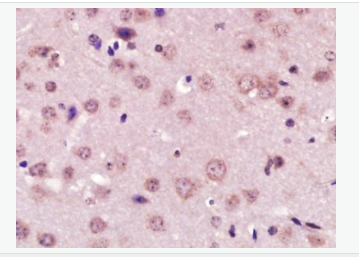
| 產品圖片 | 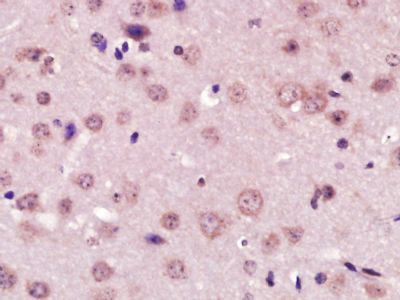 Paraformaldehyde-fixed, paraffin embedded (Mouse brain); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (XPC) Polyclonal Antibody, Unconjugated (bs-6634R) at 1:400 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. Paraformaldehyde-fixed, paraffin embedded (Mouse brain); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (XPC) Polyclonal Antibody, Unconjugated (bs-6634R) at 1:400 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining.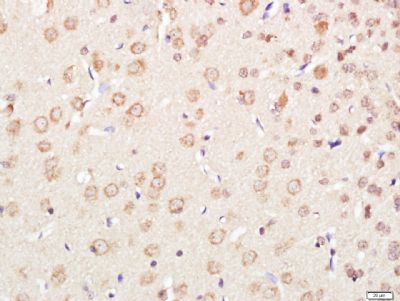 Paraformaldehyde-fixed, paraffin embedded (rat brain); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (XPC) Polyclonal Antibody, Unconjugated (bs-6634R) at 1:200 overnight at 4°C, followed by a conjugated secondary (sp-0023) for 20 minutes and DAB staining. Paraformaldehyde-fixed, paraffin embedded (rat brain); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (XPC) Polyclonal Antibody, Unconjugated (bs-6634R) at 1:200 overnight at 4°C, followed by a conjugated secondary (sp-0023) for 20 minutes and DAB staining.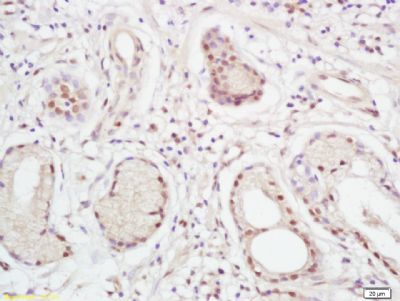 Tissue/cell: human gastric cancer; 4% Paraformaldehyde-fixed and paraffin-embedded; Tissue/cell: human gastric cancer; 4% Paraformaldehyde-fixed and paraffin-embedded;Antigen retrieval: citrate buffer ( 0.01M, pH 6.0 ), Boiling bathing for 15min; Block endogenous peroxidase by 3% Hydrogen peroxide for 30min; Blocking buffer (normal goat serum,C-0005) at 37℃ for 20 min; Incubation: Anti-XPC Polyclonal Antibody, Unconjugated(bs-6634R) 1:200, overnight at 4°C, followed by conjugation to the secondary antibody(SP-0023) and DAB(C-0010) staining 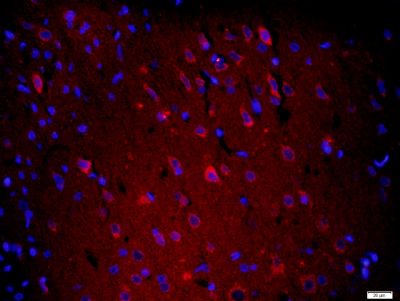 Paraformaldehyde-fixed, paraffin embedded (rat brain); Antigen retrieval by boiling in sodium citrate buffer (pH6) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (XPC) Polyclonal Antibody, Unconjugated (bs-6634R) at 1:200 overnight at 4°C, followed by a conjugated secondary (bs-0295G-Cy3) at [1:500] for 90 minutes and DAPI staining of the nuclei. Paraformaldehyde-fixed, paraffin embedded (rat brain); Antigen retrieval by boiling in sodium citrate buffer (pH6) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (XPC) Polyclonal Antibody, Unconjugated (bs-6634R) at 1:200 overnight at 4°C, followed by a conjugated secondary (bs-0295G-Cy3) at [1:500] for 90 minutes and DAPI staining of the nuclei. |
我要詢價
*聯系方式:
(可以是QQ、MSN、電子郵箱、電話等,您的聯系方式不會被公開)
*內容: